Training & evaluation with the built-in methods
Source:vignettes/training_with_built_in_methods.Rmd
training_with_built_in_methods.RmdIntroduction
This guide covers training, evaluation, and prediction (inference)
models when using built-in APIs for training & validation (such as
fit(), evaluate() and
predict()).
If you are interested in leveraging fit() while
specifying your own training step function, see the Customizing what happens in
fit() guide.
If you are interested in writing your own training & evaluation loops from scratch, see the guide Writing a training loop from scratch.
In general, whether you are using built-in loops or writing your own, model training & evaluation works strictly in the same way across every kind of Keras model – Sequential models, models built with the Functional API, and models written from scratch via model subclassing.
API overview: a first end-to-end example
When passing data to the built-in training loops of a model, you should either use:
- Arrays (if your data is small and fits in memory)
-
tf_datasetobjects - PyTorch
DataLoaderinstances
In the next few paragraphs, we’ll use the MNIST dataset as NumPy arrays, in order to demonstrate how to use optimizers, losses, and metrics. Afterwards, we’ll take a close look at each of the other options.
Let’s consider the following model (here, we build in with the Functional API, but it could be a Sequential model or a subclassed model as well):
inputs <- keras_input(shape = 784, name="digits")
outputs <- inputs |>
layer_dense(units = 64, activation = "relu", name = "dense_1") |>
layer_dense(units = 64, activation = "relu", name = "dense_2") |>
layer_dense(units = 10, activation = "softmax", name = "predictions")
model <- keras_model(inputs = inputs, outputs = outputs)
summary(model)## Model: "functional"
## ┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━┓
## ┃ Layer (type) ┃ Output Shape ┃ Param # ┃
## ┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━┩
## │ digits (InputLayer) │ (None, 784) │ 0 │
## ├─────────────────────────────────┼────────────────────────┼───────────────┤
## │ dense_1 (Dense) │ (None, 64) │ 50,240 │
## ├─────────────────────────────────┼────────────────────────┼───────────────┤
## │ dense_2 (Dense) │ (None, 64) │ 4,160 │
## ├─────────────────────────────────┼────────────────────────┼───────────────┤
## │ predictions (Dense) │ (None, 10) │ 650 │
## └─────────────────────────────────┴────────────────────────┴───────────────┘
## Total params: 55,050 (215.04 KB)
## Trainable params: 55,050 (215.04 KB)
## Non-trainable params: 0 (0.00 B)Here’s what the typical end-to-end workflow looks like, consisting of:
- Training
- Validation on a holdout set generated from the original training data
- Evaluation on the test data
We’ll use MNIST data for this example.
c(c(x_train, y_train), c(x_test, y_test)) %<-% dataset_mnist()
# Preprocess the data (these are NumPy arrays)
x_train <- array_reshape(x_train, c(60000, 784)) / 255
x_test <- array_reshape(x_test, c(10000, 784)) / 255
# Reserve 10,000 samples for validation
x_val <- x_train[1:10000,]
y_val <- y_train[1:10000]
x_train <- x_train[-c(1:10000),]
y_train <- y_train[-c(1:10000)]We specify the training configuration (optimizer, loss, metrics):
model |> compile(
# Optimizer
optimizer = optimizer_rmsprop(),
# Loss function to minimize
loss = loss_sparse_categorical_crossentropy(),
# List of metrics to monitor
metrics = list(metric_sparse_categorical_accuracy())
)We call fit(), which will train the model by slicing the
data into “batches” of size batch_size, and repeatedly
iterating over the entire dataset for a given number of
epochs.
history <- model |> fit(
x_train, y_train,
batch_size = 64,
epochs = 2,
# We pass some validation for
# monitoring validation loss and metrics
# at the end of each epoch
validation_data = list(x_val, y_val)
)## Epoch 1/2
## 782/782 - 3s - 3ms/step - loss: 0.3410 - sparse_categorical_accuracy: 0.9034 - val_loss: 0.1855 - val_sparse_categorical_accuracy: 0.9460
## Epoch 2/2
## 782/782 - 1s - 1ms/step - loss: 0.1590 - sparse_categorical_accuracy: 0.9538 - val_loss: 0.1323 - val_sparse_categorical_accuracy: 0.9621The returned history object holds a record of the loss
values and metric values during training:
history##
## Final epoch (plot to see history):
## loss: 0.159
## sparse_categorical_accuracy: 0.9538
## val_loss: 0.1323
## val_sparse_categorical_accuracy: 0.9621We evaluate the model on the test data via
evaluate():
# Evaluate the model on the test data using `evaluate`
results <- model |> evaluate(x_test, y_test, batch_size=128)## 79/79 - 0s - 4ms/step - loss: 0.1273 - sparse_categorical_accuracy: 0.9621
str(results)## List of 2
## $ loss : num 0.127
## $ sparse_categorical_accuracy: num 0.962
# Generate predictions (probabilities -- the output of the last layer)
# on new data using `predict`
predictions <- model |> predict(x_test[1:2,])## 1/1 - 0s - 159ms/step
dim(predictions)## [1] 2 10Now, let’s review each piece of this workflow in detail.
The compile() method: specifying a loss, metrics, and
an optimizer
To train a model with fit(), you need to specify a loss
function, an optimizer, and optionally, some metrics to monitor.
You pass these to the model as arguments to the
compile() method:
model |> compile(
optimizer = optimizer_rmsprop(learning_rate = 1e-3),
loss = loss_sparse_categorical_crossentropy(),
metrics = list(metric_sparse_categorical_accuracy())
)The metrics argument should be a list – your model can
have any number of metrics.
If your model has multiple outputs, you can specify different losses and metrics for each output, and you can modulate the contribution of each output to the total loss of the model. You will find more details about this in the Passing data to multi-input, multi-output models section.
Note that if you’re satisfied with the default settings, in many cases the optimizer, loss, and metrics can be specified via string identifiers as a shortcut:
model |> compile(
optimizer = "rmsprop",
loss = "sparse_categorical_crossentropy",
metrics = c("sparse_categorical_accuracy")
)For later reuse, let’s put our model definition and compile step in functions; we will call them several times across different examples in this guide.
get_uncompiled_model <- function() {
inputs <- keras_input(shape = 784, name = "digits")
outputs <- inputs |>
layer_dense(units = 64, activation = "relu", name = "dense_1") |>
layer_dense(units = 64, activation = "relu", name = "dense_2") |>
layer_dense(units = 10, activation = "softmax", name = "predictions")
keras_model(inputs = inputs, outputs = outputs)
}
get_compiled_model <- function() {
model <- get_uncompiled_model()
model |> compile(
optimizer = "rmsprop",
loss = "sparse_categorical_crossentropy",
metrics = c("sparse_categorical_accuracy")
)
model
}Many built-in optimizers, losses, and metrics are available
In general, you won’t have to create your own losses, metrics, or optimizers from scratch, because what you need is likely to be already part of the Keras API:
Optimizers:
- [
optimizer_sgd()] (with or without momentum) - [
optimizer_rmsprop()] - [
optimizer_adam()] - etc.
Losses:
Metrics:
- [
metric_auc()] - [
metric_precision()] - [
metric_recall()] - etc.
Custom losses
If you need to create a custom loss, Keras provides three ways to do so.
The first method involves creating a function that accepts inputs
y_true and y_pred. The following example shows
a loss function that computes the mean squared error between the real
data and the predictions:
custom_mean_squared_error <- function(y_true, y_pred) {
op_mean(op_square(y_true - y_pred), axis = -1)
}
model <- get_uncompiled_model()
model |> compile(optimizer = "adam",
loss = custom_mean_squared_error)
# We need to one-hot encode the labels to use MSE
y_train_one_hot <- op_one_hot(y_train, num_classes = 10)
model |> fit(x_train, y_train_one_hot, batch_size = 64, epochs = 2)## Epoch 1/2
## 782/782 - 2s - 2ms/step - loss: 0.0157
## Epoch 2/2
## 782/782 - 1s - 1ms/step - loss: 0.0074If you need a loss function that takes in parameters beside
y_true and y_pred, you can subclass the Keras
base Loss class using [Loss()] and implement
the following two methods:
-
initialize(): accept parameters to pass during the call of your loss function -
call(y_true, y_pred): use the targets (y_true) and the model predictions (y_pred) to compute the model’s loss
Let’s say you want to use mean squared error, but with an added term that will de-incentivize prediction values far from 0.5 (we assume that the categorical targets are one-hot encoded and take values between 0 and 1). This creates an incentive for the model not to be too confident, which may help reduce overfitting (we won’t know if it works until we try!).
Here’s how you would do it:
loss_custom_mse <- Loss(
classname = "CustomMSE",
initialize = function(regularization_factor = 0.1, name = "custom_mse") {
super$initialize(name = name)
self$regularization_factor <- regularization_factor
},
call = function(y_true, y_pred) {
mse <- op_mean(op_square(y_true - y_pred), axis = -1)
reg <- op_mean(op_square(0.5 - y_pred), axis = -1)
mse + reg * self$regularization_factor
}
)
model <- get_uncompiled_model()
model |> compile(optimizer="adam", loss = loss_custom_mse())
y_train_one_hot <- op_one_hot(y_train, num_classes=10)
model |> fit(x_train, y_train_one_hot, batch_size=64, epochs=1)## 782/782 - 2s - 2ms/step - loss: 0.0388Custom metrics
If you need a metric that isn’t part of the API, you can easily
create custom metrics by subclassing the Keras base Metric
class using [Metric()]. You will need to implement 4
methods:
-
initialize(), in which you will create state variables for your metric. -
update_state(y_true, y_pred, sample_weight = NULL), which uses the targets y_true and the model predictions y_pred to update the state variables. -
result(), which uses the state variables to compute the final results. -
reset_state(), which reinitializes the state of the metric.
State update and results computation are kept separate (in
update_state() and result(), respectively)
because in some cases, the results computation might be very expensive
and would only be done periodically.
Here’s a simple example showing how to implement a
CategoricalTruePositives metric that counts how many
samples were correctly classified as belonging to a given class:
metric_categorical_true_positives <- Metric(
"CategoricalTruePositives",
initialize = function(name = "categorical_true_positives", ...) {
super$initialize(name = name, ...)
self$true_positives <- self$add_variable(shape = shape(),
name = "ctp",
initializer = "zeros")
},
update_state = function(y_true, y_pred, sample_weight = NULL) {
y_pred <- op_argmax(y_pred, axis = 2) |> op_reshape(c(-1, 1))
values <- op_cast(y_true, "int32") == op_cast(y_pred, "int32")
values <- op_cast(values, "float32")
if (!is.null(sample_weight)) {
sample_weight <- op_cast(sample_weight, "float32")
values <- op_multiply(values, sample_weight)
}
self$true_positives$assign_add(op_sum(values))
},
result = function() {
self$true_positives$value
},
reset_state = function() {
self$true_positives$assign(0.0)
}
)
model <- get_uncompiled_model()
model |> compile(
optimizer = optimizer_rmsprop(learning_rate = 1e-3),
loss = loss_sparse_categorical_crossentropy(),
metrics = c(metric_categorical_true_positives())
)
history <- model |> fit(x_train, y_train, batch_size = 64, epochs = 3)## Epoch 1/3
## 782/782 - 2s - 2ms/step - categorical_true_positives: 285017.0000 - loss: 0.3444
## Epoch 2/3
## 782/782 - 1s - 1ms/step - categorical_true_positives: 284268.0000 - loss: 0.1658
## Epoch 3/3
## 782/782 - 1s - 1ms/step - categorical_true_positives: 284173.0000 - loss: 0.1207Handling losses and metrics that don’t fit the standard signature
The overwhelming majority of losses and metrics can be computed from
y_true and y_pred, where y_pred
is an output of your model – but not all of them. For instance, a
regularization loss may only require the activation of a layer (there
are no targets in this case), and this activation may not be a model
output.
In such cases, you can call self$add_loss(loss_value)
from inside the call method of a custom layer. Losses added in this way
get added to the “main” loss during training (the one passed to
compile()). Here’s a simple example that adds activity
regularization (note that activity regularization is built-in in all
Keras layers – this layer is just for the sake of providing a concrete
example):
layer_custom_activity_regularizer <- Layer(
"ActivityRegularization",
call = function(inputs) {
self$add_loss(op_sum(inputs) * 0.1)
inputs # Pass-through layer.
}
)
inputs <- keras_input(shape = 784, name = "digits")
outputs <- inputs |>
layer_dense(units = 32, activation = "relu", name = "dense_1") |>
layer_custom_activity_regularizer() |>
layer_dense(units = 64, activation = "relu", name = "dense_2") |>
layer_dense(units = 10, name = "predictions")
model <- keras_model(inputs = inputs, outputs = outputs)
model |> compile(optimizer = optimizer_rmsprop(learning_rate = 1e-3),
loss = loss_sparse_categorical_crossentropy(from_logits = TRUE))
# The displayed loss will be much higher than before
# due to the regularization component.
model |> fit(x_train, y_train, batch_size = 64, epochs = 1)## 782/782 - 2s - 2ms/step - loss: 2.3721Note that when you pass losses via add_loss(), it
becomes possible to call compile() without a loss function,
since the model already has a loss to minimize.
Consider the following LogisticEndpoint layer: it takes
as inputs targets & logits, and it tracks a crossentropy loss via
add_loss().
layer_logistic_endpoint <- Layer(
"LogisticEndpoint",
initialize = function(name = NULL) {
super$initialize(name = name)
self$loss_fn <- loss_binary_crossentropy(from_logits = TRUE)
},
call = function(targets, logits, sample_weights = NULL) {
# Compute the training-time loss value and add it
# to the layer using `self.add_loss()`.
loss <- self$loss_fn(targets, logits, sample_weights)
self$add_loss(loss)
# Return the inference-time prediction tensor (for `predict()`).
op_softmax(logits)
}
)You can use it in a model with two inputs (input data & targets),
compiled without a loss argument, like this:
inputs <- keras_input(shape = 3, name = "inputs")
targets <- keras_input(shape = 10, name = "targets")
logits <- inputs |> layer_dense(10)
predictions <- layer_logistic_endpoint(name = "predictions")(targets, logits)
model <- keras_model(inputs = list(inputs, targets),
outputs = predictions)
model |> compile(optimizer = "adam") # No loss argument!
data <- list(
inputs = random_normal(c(3, 3)),
targets = random_normal(c(3, 10))
)
model |> fit(data, epochs = 1)## 1/1 - 0s - 437ms/step - loss: 0.9638For more information about training multi-input models, see the section Passing data to multi-input, multi-output models.
Automatically setting apart a validation holdout set
In the first end-to-end example you saw, we used the
validation_data argument to pass a list of arrays
list(x_val, y_val) to the model for evaluating a validation
loss and validation metrics at the end of each epoch.
Here’s another option: the argument validation_split
allows you to automatically reserve part of your training data for
validation. The argument value represents the fraction of the data to be
reserved for validation, so it should be set to a number higher than 0
and lower than 1. For instance, validation_split = 0.2
means “use 20% of the data for validation”, and
validation_split = 0.6 means “use 60% of the data for
validation”.
The way the validation is computed is by taking the last x% samples
of the arrays received by the fit() call, before any
shuffling.
Note that you can only use validation_split when
training with NumPy data.
model <- get_compiled_model()
model |> fit(x_train, y_train,
batch_size = 64,
validation_split = 0.2, epochs = 1)## 625/625 - 2s - 3ms/step - loss: 0.3817 - sparse_categorical_accuracy: 0.8919 - val_loss: 0.1953 - val_sparse_categorical_accuracy: 0.9431Training & evaluation using TF Dataset objects
In the past few paragraphs, you’ve seen how to handle losses,
metrics, and optimizers, and you’ve seen how to use the
validation_data and validation_split arguments
in fit(), when your data is passed as arrays.
Another option is to use an iterator-like, such as a
tf.data.Dataset, a PyTorch DataLoader, or an R
generator function. Let’s take look at the former.
The tfdatasets R package containes a set of utilities
for loading and preprocessing data in a way that’s fast and scalable.
For a complete guide about creating Datasets, see the tf.data
documentation.
You can use tf.data to train your Keras models
regardless of the backend you’re using – whether it’s JAX, PyTorch, or
TensorFlow. You can pass a Dataset instance
directly to the methods fit(), evaluate(), and
predict():
library(tfdatasets, exclude = "shape")
model <- get_compiled_model()
# First, let's create a training Dataset instance.
# For the sake of our example, we'll use the same MNIST data as before.
train_dataset <- tensor_slices_dataset(list(x_train, y_train))
# Shuffle and slice the dataset.
train_dataset <- train_dataset |>
dataset_shuffle(buffer_size=1024) |>
dataset_batch(64)
# Now we get a test dataset.
test_dataset <-
tensor_slices_dataset(list(x_test, y_test)) |>
dataset_batch(64)
# Since the dataset already takes care of batching,
# we don't pass a `batch_size` argument.
model |> fit(train_dataset, epochs = 3)## Epoch 1/3
## 782/782 - 2s - 2ms/step - loss: 0.3365 - sparse_categorical_accuracy: 0.9041
## Epoch 2/3
## 782/782 - 1s - 1ms/step - loss: 0.1606 - sparse_categorical_accuracy: 0.9521
## Epoch 3/3
## 782/782 - 1s - 1ms/step - loss: 0.1178 - sparse_categorical_accuracy: 0.9646
# You can also evaluate or predict on a dataset.
result <- model |> evaluate(test_dataset)## 157/157 - 1s - 4ms/step - loss: 0.1213 - sparse_categorical_accuracy: 0.9596
result## $loss
## [1] 0.1213005
##
## $sparse_categorical_accuracy
## [1] 0.9596Note that the Dataset is reset at the end of each epoch,
so it can be reused of the next epoch.
If you want to run training only on a specific number of batches from
this Dataset, you can pass the steps_per_epoch argument,
which specifies how many training steps the model should run using this
Dataset before moving on to the next epoch.
model <- get_compiled_model()
# Prepare the training dataset
train_dataset <- tensor_slices_dataset(list(x_train, y_train))
train_dataset <- train_dataset |>
dataset_shuffle(buffer_size = 1024) |>
dataset_batch(64)
# Only use the 100 batches per epoch (that's 64 * 100 samples)
model |> fit(train_dataset, epochs = 3, steps_per_epoch = 100)## Epoch 1/3
## 100/100 - 1s - 8ms/step - loss: 0.8017 - sparse_categorical_accuracy: 0.7806
## Epoch 2/3
## 100/100 - 0s - 1ms/step - loss: 0.3661 - sparse_categorical_accuracy: 0.9006
## Epoch 3/3
## 100/100 - 0s - 1ms/step - loss: 0.3009 - sparse_categorical_accuracy: 0.9106You can also pass a Dataset instance as the
validation_data argument in fit():
model <- get_compiled_model()
# Prepare the training dataset
train_dataset <- tensor_slices_dataset(list(x_train, y_train))
train_dataset <- train_dataset |>
dataset_shuffle(buffer_size=1024) |>
dataset_batch(64)
# Prepare the validation dataset
val_dataset <- tensor_slices_dataset(list(x_val, y_val))
val_dataset <- val_dataset |> dataset_batch(64)
model |> fit(train_dataset, epochs = 1, validation_data = val_dataset)## 782/782 - 2s - 3ms/step - loss: 0.3428 - sparse_categorical_accuracy: 0.9022 - val_loss: 0.2337 - val_sparse_categorical_accuracy: 0.9291At the end of each epoch, the model will iterate over the validation dataset and compute the validation loss and validation metrics.
If you want to run validation only on a specific number of batches
from this dataset, you can pass the validation_steps
argument, which specifies how many validation steps the model should run
with the validation dataset before interrupting validation and moving on
to the next epoch:
model <- get_compiled_model()
# Prepare the training dataset
train_dataset <- tensor_slices_dataset(list(x_train, y_train))
train_dataset <- train_dataset |>
dataset_shuffle(buffer_size = 1024) |>
dataset_batch(64)
# Prepare the validation dataset
val_dataset <- tensor_slices_dataset(list(x_val, y_val))
val_dataset <- val_dataset |> dataset_batch(64)
model %>% fit(
train_dataset,
epochs = 1,
# Only run validation using the first 10 batches of the dataset
# using the `validation_steps` argument
validation_data = val_dataset,
validation_steps = 10,
)## 782/782 - 2s - 3ms/step - loss: 0.3391 - sparse_categorical_accuracy: 0.9035 - val_loss: 0.1997 - val_sparse_categorical_accuracy: 0.9391Note that the validation dataset will be reset after each use (so that you will always be evaluating on the same samples from epoch to epoch).
The argument validation_split (generating a holdout set
from the training data) is not supported when training from
Dataset objects, since this feature requires the ability to
index the samples of the datasets, which is not possible in general with
the Dataset API.
Using sample weighting and class weighting
With the default settings the weight of a sample is decided by its frequency in the dataset. There are two methods to weight the data, independent of sample frequency:
- Class weights
- Sample weights
Class weights
This is set by passing a named list to the class_weight
argument to fit(). This list maps class indices to the
weight that should be used for samples belonging to this class.
This can be used to balance classes without resampling, or to train a model that gives more importance to a particular class.
For instance, if class “0” is half as represented as class “1” in
your data, you could use
model |> fit(..., class_weight = c("0" = 1, "1" = 0.5)).
Here’s an R example where we use class weights or sample weights to give more importance to the correct classification of class #5 (which is the digit “5” in the MNIST dataset).
class_weight <- c(
"0" = 1.0,
"1" = 1.0,
"2" = 1.0,
"3" = 1.0,
"4" = 1.0,
# Set weight "2" for class "5",
# making this class 2x more important
"5" = 2.0,
"6" = 1.0,
"7" = 1.0,
"8" = 1.0,
"9" = 1.0
)
model <- get_compiled_model()
model |> fit(x_train, y_train,
class_weight = class_weight,
batch_size = 64, epochs = 1)## 782/782 - 2s - 2ms/step - loss: 0.3713 - sparse_categorical_accuracy: 0.9018Sample weights
For fine grained control, or if you are not building a classifier,
you can use sample_weights.
- When training from R arrays: Pass the
sample_weightargument tofit(). - When training from
tf_datasetor any other sort of iterator: yield(input_batch, label_batch, sample_weight_batch)tuples.
A “sample weights” array is an array of numbers that specify how much weight each sample in a batch should have in computing the total loss. It is commonly used in imbalanced classification problems (the idea being to give more weight to rarely-seen classes).
When the weights used are ones and zeros, the array can be used as a mask for the loss function (entirely discarding the contribution of certain samples to the total loss).
sample_weight <- rep(1.0, length(y_train))
sample_weight[y_train == 5] <- 2.0
model <- get_compiled_model()
model |> fit(
x_train, y_train,
sample_weight = sample_weight,
batch_size = 64, epochs = 1
)## 782/782 - 2s - 2ms/step - loss: 0.3740 - sparse_categorical_accuracy: 0.9015Here’s a matching Dataset example:
sample_weight <- rep(1.0, length(y_train))
sample_weight[y_train == 5] <- 2.0
# Create a Dataset that includes sample weights
# (3rd element in the return tuple).
train_dataset <- tensor_slices_dataset(list(
x_train, y_train, sample_weight
))
# Shuffle and slice the dataset.
train_dataset <- train_dataset |>
dataset_shuffle(buffer_size = 1024) |>
dataset_batch(64)
model <- get_compiled_model()
model |> fit(train_dataset, epochs = 1)## 782/782 - 2s - 2ms/step - loss: 0.3654 - sparse_categorical_accuracy: 0.9057Passing data to multi-input, multi-output models
In the previous examples, we were considering a model with a single
input (a tensor of shape (764)) and a single output (a
prediction tensor of shape (10)). But what about models
that have multiple inputs or outputs?
Consider the following model, which has an image input of shape
(32, 32, 3) (that’s (height, width, channels))
and a time series input of shape (NA, 10) (that’s
(timesteps, features)). Our model will have two outputs
computed from the combination of these inputs: a “score” (of shape
(1)) and a probability distribution over five classes (of
shape (5)).
image_input <- keras_input(c(32, 32, 3), name = "img_input")
timeseries_input <- keras_input(c(NA, 10), name = "ts_input")
x1 <- image_input |>
layer_conv_2d(filters = 3, kernel_size = c(3, 3)) |>
layer_global_max_pooling_2d()
x2 <- timeseries_input |>
layer_conv_1d(filters = 3, kernel_size = 3) |>
layer_global_max_pooling_1d()
x <- layer_concatenate(x1, x2)
score_output <- layer_dense(x, 1, name = "score_output")
class_output <- layer_dense(x, 5, name = "class_output")
model <- keras_model(
inputs = list(image_input, timeseries_input),
outputs = list(score_output, class_output)
)Let’s plot this model, so you can clearly see what we’re doing here (note that the shapes shown in the plot are batch shapes, rather than per-sample shapes).
plot(model, show_shapes = TRUE)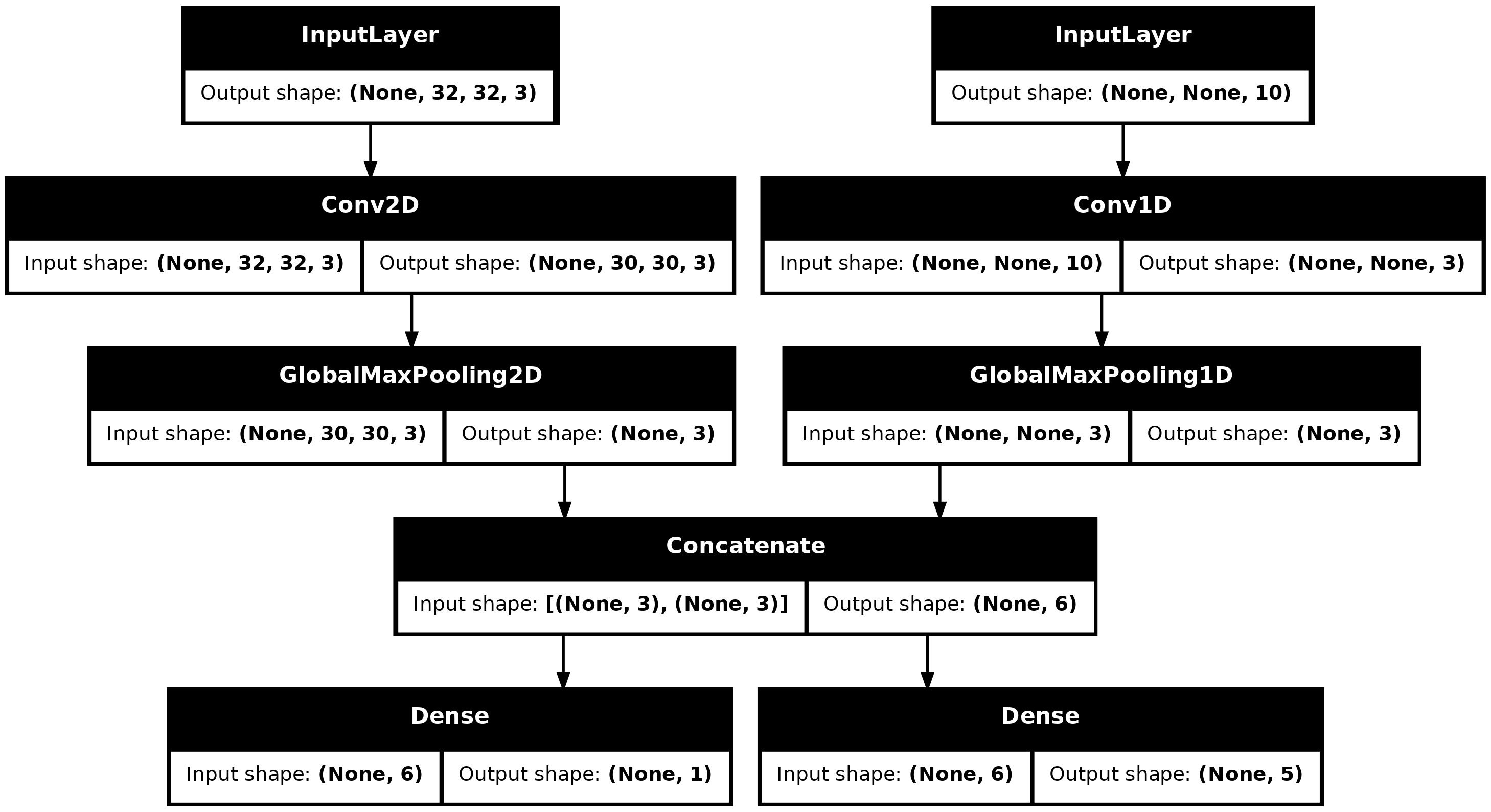
plot of chunk unnamed-chunk-26
At compilation time, we can specify different losses to different outputs, by passing the loss functions as a list:
model |> compile(
optimizer = optimizer_rmsprop(1e-3),
loss = list(
loss_mean_squared_error(),
loss_categorical_crossentropy()
)
)If we only passed a single loss function to the model, the same loss function would be applied to every output (which is not appropriate here).
Likewise for metrics:
model |> compile(
optimizer = optimizer_rmsprop(1e-3),
loss = list(
loss_mean_squared_error(),
loss_categorical_crossentropy()
),
metrics = list(
list(
metric_mean_absolute_error(),
metric_mean_absolute_percentage_error()
),
list(metric_categorical_accuracy())
)
)Since we gave names to our output layers, we could also specify per-output losses and metrics via a named list:
model |> compile(
optimizer = optimizer_rmsprop(1e-3),
loss = list(
score_output = loss_mean_squared_error(),
class_output = loss_categorical_crossentropy()
),
metrics = list(
score_output = list(
metric_mean_absolute_error(),
metric_mean_absolute_percentage_error()
),
class_output = list(metric_categorical_accuracy())
)
)We recommend the use of names if you have more than 2 outputs.
It’s possible to give different weights to different output-specific
losses (for instance, one might wish to privilege the “score” loss in
our example, by giving to 2x the importance of the class loss), using
the loss_weights argument:
model |> compile(
optimizer = optimizer_rmsprop(1e-3),
loss = list(
score_output = loss_mean_squared_error(),
class_output = loss_categorical_crossentropy()
),
metrics = list(
score_output = list(
metric_mean_absolute_error(),
metric_mean_absolute_percentage_error()
),
class_output = list(metric_categorical_accuracy())
),
loss_weights = list(score_output = 2.0, class_output = 1.0)
)You could also choose not to compute a loss for certain outputs, if these outputs are meant for prediction but not for training:
# loss list, positional version
model |> compile(
optimizer = optimizer_rmsprop(1e-3),
loss = list(NULL, loss_categorical_crossentropy())
)
# Or loss list, named version
model |> compile(
optimizer = optimizer_rmsprop(1e-3),
loss = list(class_output = loss_categorical_crossentropy())
)Passing data to a multi-input or multi-output model in
fit() works in a similar way as specifying a loss function
in compile: you can pass lists of arrays (with 1:1
mapping to the outputs that received a loss function) or dicts
mapping output names to arrays.
model |> compile(
optimizer = optimizer_rmsprop(1e-3),
loss = list(
loss_mean_squared_error(),
loss_binary_crossentropy()
)
)
# Generate dummy data
img_data <- random_normal(c(100, 32, 32, 3))
ts_data <- random_normal(c(100, 20, 10))
score_targets <- random_normal(c(100, 1))
class_targets <- random_normal(c(100, 5))
# Fit on unnamed lists (positional matching)
model |> fit(
list(img_data, ts_data),
list(score_targets, class_targets),
batch_size=32,
epochs=1
)## 4/4 - 2s - 540ms/step - class_output_categorical_accuracy: 0.2300 - class_output_loss: 1.8162 - loss: 3.8764 - score_output_loss: 1.6820 - score_output_mean_absolute_error: 0.8826 - score_output_mean_absolute_percentage_error: 1826.5676
# Alternatively, fit on named lists (names matching)
model <- keras_model(
inputs = list(image_input, timeseries_input),
outputs = list(score_output = score_output,
class_output = class_output)
) |> compile(
optimizer = optimizer_rmsprop(1e-3),
loss = list(
loss_mean_squared_error(),
loss_binary_crossentropy()
)
)
model |> fit(
list(img_input = img_data, ts_input = ts_data),
list(score_output = score_targets, class_output = class_targets),
batch_size = 32,
epochs = 1
)## 4/4 - 2s - 400ms/step - class_output_loss: 4.1489 - loss: 4.5443 - score_output_loss: -4.1125e-01Here’s the Dataset use case: similarly as what we did
for R arrays, the Dataset should return a tuple of named
lists (dicts).
train_dataset <- tensor_slices_dataset(list(
list(img_input = img_data, ts_input = ts_data),
list(score_output = score_targets, class_output = class_targets)
))
train_dataset <- train_dataset |>
dataset_shuffle(buffer_size = 1024) |>
dataset_batch(64)
model |> fit(train_dataset, epochs = 1)## 2/2 - 1s - 681ms/step - class_output_loss: 3.8547 - loss: 3.5725 - score_output_loss: -1.5209e-01Using callbacks
Callbacks in Keras are objects that are called at different points during training (at the start of an epoch, at the end of a batch, at the end of an epoch, etc.). They can be used to implement certain behaviors, such as:
- Doing validation at different points during training (beyond the built-in per-epoch validation)
- Checkpointing the model at regular intervals or when it exceeds a certain accuracy threshold
- Changing the learning rate of the model when training seems to be plateauing
- Doing fine-tuning of the top layers when training seems to be plateauing
- Sending email or instant message notifications when training ends or where a certain performance threshold is exceeded
- Etc.
Callbacks can be passed as a list to your call to
fit():
model <- get_compiled_model()
callbacks <- list(
callback_early_stopping(
# Stop training when `val_loss` is no longer improving
monitor = "val_loss",
# "no longer improving" being defined as "no better than 1e-2 less"
min_delta = 1e-2,
# "no longer improving" being further defined as "for at least 2 epochs"
patience = 2,
verbose = 1
)
)
model |> fit(
x_train,
y_train,
epochs = 20,
batch_size = 64,
callbacks = callbacks,
validation_split = 0.2,
)## Epoch 1/20
## 625/625 - 2s - 3ms/step - loss: 0.3695 - sparse_categorical_accuracy: 0.8961 - val_loss: 0.1873 - val_sparse_categorical_accuracy: 0.9469
## Epoch 2/20
## 625/625 - 1s - 1ms/step - loss: 0.1752 - sparse_categorical_accuracy: 0.9488 - val_loss: 0.1405 - val_sparse_categorical_accuracy: 0.9576
## Epoch 3/20
## 625/625 - 1s - 1ms/step - loss: 0.1277 - sparse_categorical_accuracy: 0.9626 - val_loss: 0.1217 - val_sparse_categorical_accuracy: 0.9644
## Epoch 4/20
## 625/625 - 1s - 1ms/step - loss: 0.1009 - sparse_categorical_accuracy: 0.9702 - val_loss: 0.1138 - val_sparse_categorical_accuracy: 0.9667
## Epoch 5/20
## 625/625 - 1s - 1ms/step - loss: 0.0823 - sparse_categorical_accuracy: 0.9758 - val_loss: 0.1086 - val_sparse_categorical_accuracy: 0.9679
## Epoch 6/20
## 625/625 - 1s - 1ms/step - loss: 0.0682 - sparse_categorical_accuracy: 0.9800 - val_loss: 0.1084 - val_sparse_categorical_accuracy: 0.9685
## Epoch 7/20
## 625/625 - 1s - 1ms/step - loss: 0.0573 - sparse_categorical_accuracy: 0.9834 - val_loss: 0.1100 - val_sparse_categorical_accuracy: 0.9698
## Epoch 8/20
## 625/625 - 1s - 1ms/step - loss: 0.0485 - sparse_categorical_accuracy: 0.9868 - val_loss: 0.1113 - val_sparse_categorical_accuracy: 0.9702
## Epoch 8: early stoppingMany built-in callbacks are available
There are many built-in callbacks already available in Keras, such as:
-
callback_model_checkpoint(): Periodically save the model. -
callback_early_stopping(): Stop training when training is no longer improving the validation metrics. -
callback_tensorboard(): periodically write model logs that can be visualized in TensorBoard (more details in the section “Visualization”). -
callback_csv_logger(): streams loss and metrics data to a CSV file. - etc.
See the callbacks documentation for the complete list.
Writing your own callback
You can create a custom callback by subclassing the base
[Callback()] class. A callback has access to its associated
model through the class property self$model.
Make sure to read the complete guide to writing custom callbacks.
Here’s a simple example saving a list of per-batch loss values during training:
callback_loss_history <- Callback(
classname = "LossHistory",
initialize = function(file = "per_training_batch_losses.txt", ...) {
super$initialize(...)
private$file <- file
},
on_train_begin = function(logs = NULL) {
private$per_batch_losses <- fastmap::faststack()
},
on_train_batch_begin = function(batch, logs = NULL) {
private$per_batch_losses$push(logs$loss)
},
on_train_end = function(logs = NULL) {
per_batch_losses <- private$per_batch_losses$as_list() |> as.numeric()
write(per_batch_losses, private$file)
}
)Checkpointing models
When you’re training model on relatively large datasets, it’s crucial to save checkpoints of your model at frequent intervals.
The easiest way to achieve this is with
[callback_model_checkpoint()]:
model <- get_compiled_model()
callbacks <- list(
callback_model_checkpoint(
# Path where to save the model
# The two parameters below mean that we will overwrite
# the current checkpoint if and only if
# the `val_loss` score has improved.
# The saved model name will include the current epoch.
filepath = "mymodel_{epoch}.keras",
save_best_only = TRUE,
# Only save a model if `val_loss` has improved.
monitor = "val_loss",
verbose = 1
)
)
model |> fit(
x_train, y_train,
epochs = 2, batch_size = 64,
callbacks = callbacks,
validation_split = 0.2
)## Epoch 1/2
##
## Epoch 1: val_loss improved from None to 0.19488, saving model to mymodel_1.keras
##
## Epoch 1: finished saving model to mymodel_1.keras
## 625/625 - 2s - 3ms/step - loss: 0.3793 - sparse_categorical_accuracy: 0.8936 - val_loss: 0.1949 - val_sparse_categorical_accuracy: 0.9447
## Epoch 2/2
##
## Epoch 2: val_loss improved from 0.19488 to 0.14489, saving model to mymodel_2.keras
##
## Epoch 2: finished saving model to mymodel_2.keras
## 625/625 - 1s - 1ms/step - loss: 0.1777 - sparse_categorical_accuracy: 0.9475 - val_loss: 0.1449 - val_sparse_categorical_accuracy: 0.9590The ModelCheckpoint callback can be used to implement
fault-tolerance: the ability to restart training from the last saved
state of the model in case training gets randomly interrupted. Here’s a
basic example:
# Prepare a directory to store all the checkpoints.
checkpoint_dir <- "./ckpt"
fs::dir_create(checkpoint_dir)
make_or_restore_model <- function() {
# Either restore the latest (best) model, or create a fresh one
# if there is no checkpoint available.
checkpoints <- Sys.glob(file.path(checkpoint_dir, "model-loss=*.keras"))
if (length(checkpoints) > 0) {
checkpoint_losses <- sub("^model-loss=([0-9.]+)\\.keras$", "\\1",
basename(checkpoints)) |> as.numeric()
best_checkpoint <- checkpoints[which.min(checkpoint_losses)]
load_model(best_checkpoint)
} else {
get_compiled_model()
}
}
model <- make_or_restore_model()
callbacks <- list(
# This callback saves the model every 100 batches.
# We include the training loss in the saved model name.
callback_model_checkpoint(
filepath = file.path(checkpoint_dir, "model-loss={loss:.2f}.keras"),
save_freq = 100
)
)
model |> fit(x_train, y_train, epochs = 1, callbacks = callbacks)## 1563/1563 - 3s - 2ms/step - loss: 0.2932 - sparse_categorical_accuracy: 0.9145You call also write your own callback for saving and restoring models.
For a complete guide on serialization and saving, see the guide to saving and serializing Models.
Using learning rate schedules
A common pattern when training deep learning models is to gradually reduce the learning as training progresses. This is generally known as “learning rate decay”.
The learning decay schedule could be static (fixed in advance, as a function of the current epoch or the current batch index), or dynamic (responding to the current behavior of the model, in particular the validation loss).
Passing a schedule to an optimizer
You can easily use a static learning rate decay schedule by passing a
schedule object as the learning_rate argument in your
optimizer:
initial_learning_rate <- 0.1
lr_schedule <- learning_rate_schedule_exponential_decay(
initial_learning_rate, decay_steps=100000, decay_rate=0.96,
staircase=TRUE
)
optimizer <- optimizer_rmsprop(learning_rate = lr_schedule)Several built-in schedules are available:
ExponentialDecay, PiecewiseConstantDecay,
PolynomialDecay, and InverseTimeDecay.
Using callbacks to implement a dynamic learning rate schedule
A dynamic learning rate schedule (for instance, decreasing the learning rate when the validation loss is no longer improving) cannot be achieved with these schedule objects, since the optimizer does not have access to validation metrics.
However, callbacks do have access to all metrics, including
validation metrics! You can thus achieve this pattern by using a
callback that modifies the current learning rate on the optimizer. In
fact, this is even built-in as
[callback_reduce_lr_on_plateau()].
Visualizing loss and metrics during training with TensorBoard
The best way to keep an eye on your model during training is to use TensorBoard – a browser-based application that you can run locally that provides you with:
- Live plots of the loss and metrics for training and evaluation
- (optionally) Visualizations of the histograms of your layer activations
- (optionally) 3D visualizations of the embedding spaces learned by
your
layer_embedding()
If you have installed TensorFlow with pip, you should be able to launch TensorBoard from the command line:
tensorboard --logdir=/full_path_to_your_logsor from R using:
tensorflow::tensorboard(logdir = "/full_path_to_your_logs")Using the TensorBoard callback
The easiest way to use TensorBoard with a Keras model and the
fit() method is with
[callback_tensorboard()].
In the simplest case, just specify where you want the callback to write logs, and you’re good to go:
tb_callback <- callback_tensorboard(
log_dir = "/full_path_to_your_logs",
histogram_freq = 0, # How often to log histogram visualizations
embeddings_freq = 0, # How often to log embedding visualizations
update_freq = "epoch", # How often to write logs (default: once per epoch)
)For more information, see callback_tensorboard().